

M.D., Stanford University
Postdoctoral, Carnegie Institution of Washington Department of Embryology
We have known for many years that a large number of rare human diseases involve dysfunction of mitochondria, the energy-producing organelles in cells. More recently, we have learned that common disorders such as Parkinson disease, Alzheimer disease, diabetes and cancer all involve aspects of mitochondrial dysfunction. We are interested in how aberrant mitochondrial-nuclear communication contributes to these diseases.
The Bogenhagen laboratory has had a role in defining the basic mechanisms of mitochondrial DNA (mtDNA) replication, transcription and repair in animal cells. All of the proteins involved in these critically important processes, over 200 in number, are encoded by nuclear genes whose protein products are imported into mitochondria. Our work has involved efforts to map promoters for transcription of mtDNA and to characterize mitochondrial DNA polymerase g, mtRNA polymerase, the architectural transcription factor TFAM and other accessory proteins. We were among the first to purify pol g, mtRNA polymerase and transcription factor TFBM2. In collaboration with the Kisker laboratory, we determined the crystal structure of the pol g accessory factor polgB and its relationship to the EM structure of the holoenzyme. We were also the first laboratory to reconstitute base excision repair using purified mitochondrial proteins.
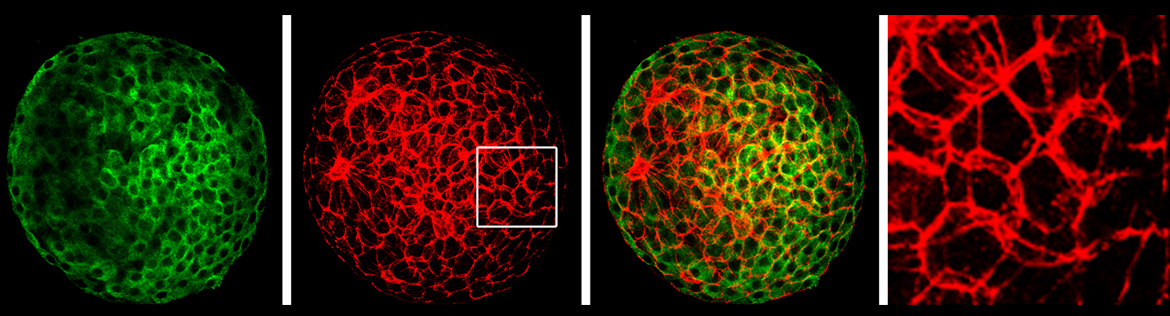
In recent years we have sought to understand the macromolecular environment in which these proteins function within mitochondria. MtDNA is not packaged in nucleosomes, but resides in tightly packed nucleoids associated with the mitochondrial inner membrane. These nucleoids are fundamental units for inheritance of mtDNA-encoded diseases. We have used super-resolution microscopy to study the structure of nucleoids and facilitated efforts to bring this innovative technology to Stony Brook University. We have used proteomics to identify proteins associated with mtDNA nucleoids. MtDNA replication, transcription, and RNA processing events are all controlled at nucleoids serving as major centers for mitochondrial biogenesis. Most recently we have expanded this work to show that the initial steps in mitochondrial ribosome biogenesis occur at the nucleoid as newly-synthesized mitochondrial ribosomal proteins first bind nascent mitochondrial ribosomal RNA while it is still being transcribed form the mtDNA. Our current research explores the coordination of nuclear and mitochondrial genetic events at nucleoids in diverse cell types in healthy and diseased states.
Recent
Lee, K.W. and Bogenhagen, D.F. (2014). Assignment of 2’-O-methyltransferases to Modification Sites on the Mammalian Mitochondrial Large Subunit 16S rRNA, J. Biol. Chem., in press.
Bogenhagen, D.F., Martin, D. and Koller, A. (2014). Initial Steps in RNA Processing and Ribosome Assembly Occur at Mitochondrial DNA Nucleoids. Cell Metabolism 19:618-629.
Lee, K.W., Okot-Kotber, C., LaComb, J.F., and Bogenhagen, D.F. (2013). Mitochondrial rRNA Methyltransferase Family Members are Positioned to Modify Nascent rRNA in Foci Near the mtDNA Nucleoid. J. Biol. Chem. 288: 31386-31399 doi:10.1074/jbc.M113.515692
Lu,. B., Lee, J., Nie, X, Li, M., Morozov, Y.I., Venkatesh, S. , Bogenhagen, D.F., Temiakov, D. and Suzuki, C. K. (2013). Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol. Cell. 49: 121-132
Brown, T.A., Tkachuk, A.N., Shtengel, G., Kopek, B.G., Bogenhagen, D.F., Hess, H.F. and. Clayton, D.A.. (2011). Super-resolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction, Mol. Cell. Biol. 31:4994-5010.
Bogenhagen, D.F. Mitochondrial DNA Nucleoid Structure. (2011) Biochim. Biophys. Acta 1819: 914-920. http://dx.doi.org/10.1016/j.bbagrm.2011.11.005
Bogenhagen, D.F. (2010) Does MtDNA Nucleoid Organization Impact Aging? Exp. Gerontology 45: 473-477.
Bogenhagen, D.F. (2009) Biochemical Isolation of mtDNA Nucleoids from Animal Cells. Methods Mol. Biol. 554: 3-14.
Zheng, L., Zhou, M., Guo, Z., Lu,H., Qian, L., Dai, H. , Qiu, J., Yakubovskaya, E., Bogenhagen, D.F., Demple, B. and Shen, B. (2008). Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell 32: 325-336. Pubmed
Liu, P., Qian, L., Sung, J.-S., deSouza-Pinto, N.C., Zheng, L., Bogenhagen, D.F., Bohr, V.A., WilsonIII, D.M., Shen, B., and Demple, B. (2008). Long-Patch Base Excision DNA Repair Dependent on FEN1 in Human Cell Mitochondria, Mol. Cell. Biol. 28:4975-4987. Pubmed
Bogenhagen, D.F. Rousseau, D. and Burke, S. (2008). The Layered Structure of Human mtDNA Nucleoids. J. Biol. Chem. 283: 3665-3675. Pubmed
Watkins, J., Basu, S. and Bogenhagen, D.F. (2008). Quantitative Proteomic Analysis of Cell Cycle, Metabolic and Cytoskeletal Changes Accompanying P19 Cell Neuronal Differentiation. J. Proteome Res. 7: 328-338. Pubmed
Yakubovskaya, E., Lukin, M., Chen, Z., Berriman, J., Wall, J.S., Kobayashi,R., Kisker, C., and Bogenhagen, D.F. (2007). The EM Structure of Human DNA Polymerase Gamma Reveals a Localized Contact between the Catalytic and Accessory Subunits. EMBO J., 26: 4283-91. Pubmed
Wang, Y. and Bogenhagen, D.F. (2006). Human mtDNA Nucleoids Are Linked to Protein Folding Machinery and Metabolic Enzymes at the Mitochondrial Inner Membrane. J. Biol. Chem., 281: 25791-25802. Pubmed
Basu, S., Bremer, E., Zhou, C. and Bogenhagen, D.F. (2006) MiGenes: A Searchable Interspecies Database of Mitochondrial Proteins Curated Using Gene Ontology Annotation. Bioinformatics 22: 485-492. Pubmed
Yakubovskaya, E., Chen, Z., Carrodeguas, J.A., Kisker, C. and Bogenhagen, D.F. (2006). Functional Human Mitochondrial DNA Polymerase Gamma Forms a Heterotrimer. J. Biol. Chem. 281: 374-382. Pubmed
Pinz, K.G. and Bogenhagen, D.F. (2006). The Influence of the DNA Polymerase Gamma Accessory Subunit on Base Excision Repair by the Catalytic Subunit. DNA Repair 6: 121-128. Pubmed
Selected Earlier Publications
Bogenhagen, DF, Wang,Y, Shen, E and Kobayashi, R. (2003). Protein Components of Mitochondrial DNA Nucleoids in Higher Eukaryotes. Molecular and Cellular Proteomics, 2: 1205-1216. PubMed
Carrodeguas JA, Pinz KG and Bogenhagen DF. (2002). DNA binding properties of human pol GammaB. J Biol Chem. 277:50008-14. PubMed
Carrodeguas, J.A., Theis, K., Bogenhagen, D.F. and Kisker, C. (2001). Crystal Structure and Deletion Analysis Show that the Accessory Subunit of Mammalian DNA Polymerase gamma, Pol gammaB, Functions as a Homodimer. Mol. Cell 7: 43-54. Pubmed
Perez-Jannotti, R, Klein, SM and Bogenhagen, DF. (2001). Two forms of mitochondrial DNA ligase III are produced in Xenopus laevis oocytes. J. Biol. Chem. 276: 48978-48987. PubMed
Shen, EL and Bogenhagen, DF. (2001). Developmentally-Regulated Packaging of Mitochondrial DNA by the HMG-box Protein mtTFA during Xenopus Oogenesis. Nucleic Acids Res. 29: 2822-2828. PubMed
Pinz, KG and Bogenhagen, DF. (2000) Characterization of a catalytically slow AP lyase activity in DNA polymerase gamma and other family A DNA polymerases. J Biol Chem. 275:12509-14. PubMed
Carrodeguas JA, Kobayashi R, Lim SE, Copeland WC and Bogenhagen DF. (1999) The accessory subunit of Xenopus laevis mitochondrial DNA polymerase gamma increases processivity of the catalytic subunit of human DNA polymerase gamma and is related to class II aminoacyl-tRNA synthetases. Mol Cell Biol. 19:4039-46. PubMed
Bogenhagen, D.F. (1999). Repair of Mitochondrial DNA. Am. J. Hum. Genet. 64:1276-1281 Pubmed
Pinz, K.G. and Bogenhagen, D.F. (1998). Efficient Repair of Abasic Sites in DNA by Mitochondrial Enzymes. Mol. Cell. Biol. 18: 1257-1265. Pubmed
daniel.bogenhagen@stonybrook.edu
631-444-3068